The end of the nineteenth century was a turning point in the field of medicine marked by incredible accomplishments, such as the discovery of the smallpox vaccine and antibiotics. Medical practitioners had since been looking towards emerging developments in biomedical research.
Biomarker research
Rather than delivering drugs that target a general population, biomarker research now aims to support the development of drugs targeted for specific populations. But biomarker discovery remains challenging due to high false discovery rates in the early stages of research - markers that seem to be clinically useful in preliminary studies often fail to differentiate patients from healthy subjects when tested out in larger populations. It is thus critical to develop technologies that can improve the discovery and translation process.
Differentiating proteins
The main technology for proteomic discovery is mass spectrometry (MS), a technology used to identify molecules based on how much they (and their constituent parts) weigh. MS-based proteomics experiments are often designed to address the question, “what are the differences between the sets of proteins expressed in normal versus diseased states?”
Differentially expressed proteins then undergo targeted experiments in larger numbers of patient samples to verify that they are still valid in larger populations. The candidates with the highest accuracy, sensitivity and specificity in identifying disease are finally selected for development and commercialization as clinical lab diagnostics.
Collect and interpret massive amount of data
Many research efforts are oriented around MS-based approaches to improving biomarker discovery. Two software tools, STRAP [7] and STRAP PTM (post translational modification) were developed to help researchers interpret their data from the discovery phase of research. STRAP and STRAP PTM provide visual aids that describe differences in protein function and modification, and use several algorithms to identify significant differences between several groups (e.g., “Healthy”, “Diseased”, and “Treated”) in the experiment.
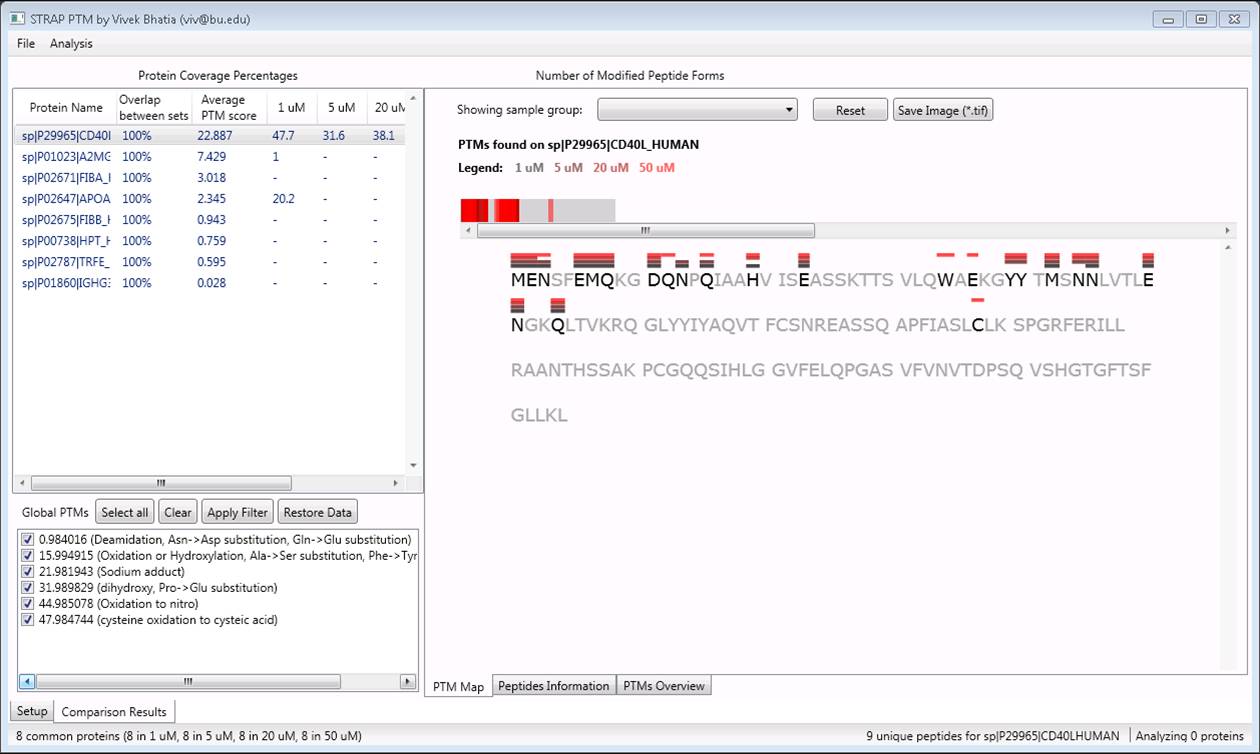
Table shown here visually summarizes an otherwise cumbersome data set as a visual of protein structure. The differently colored bars above individual amino acids indicate the various modifications observed by MS among several experimental groups.
Doing in minutes what normally may take several days, STRAP PTM rapidly helps direct researchers to interesting differences in protein structure across healthy subjects or sick patients. These differences may constitute “candidate biomarkers” that warrant a deeper investigation.
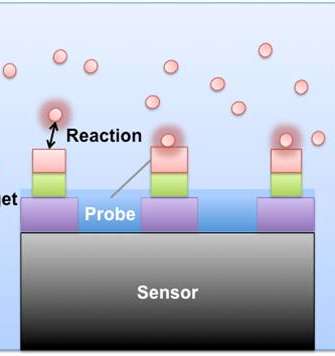
The IRIS-MALDI sensor (see featured image) uses a specially designed surface to capture several different, but specific molecules in a fluid such as blood or urine. As the potential biomarker molecules are captured on the surface, the sensor’s height increases by several nanometers at a time. After measuring the surface height optically, we can determine how much of each molecule was in the original fluid.
Using MS, we can also understand what was bound to the surface, including modified forms of the proteins. Because the sensor contains tens to hundreds of spots to capture different proteins, multiple biomarker tests can be run on a single chip.
Because this technology is cheap and relatively easy to use compared to other biomarker discovery tools, it has the potential to be used in clinical labs for diagnostic purposes. IRIS-MALDI sensors could dramatically reduce the time and resources required to develop a diagnostic assay for clinical lab use. Since most biomarker candidates never survive translation to the clinical lab, this multi-purpose use of IRIS-MALDI chips would certainly be no small feat since it aims to eliminate the gap between discovery and translation.
As physical technologies improve, massive amounts of data are being collected without an easy way to interpret them. Parallel development of software technologies to analyze these data will thus be the key to improving the power of biomarkers in medicine.
With the help of advanced biotechnology, risk stratification, diagnosis and treatment of diseases will be less expensive and more effective in the future. An easier discovery workflow built upon tools such as STRAP PTM and IRIS-MALDI sensors would strengthen the quality of preventive health care and reduce the long-term financial burden on the health care system.